Beta-Lactamases Class A Family
This is a case study that was analyzed in one of our articles where we made a link between local frustration and protein enzymes: Freiberger et al PNAS 2019.
In the following link you can find a FrustraEvo reproduction of this example:
FrustraEvo job results
Biological Interpretation:
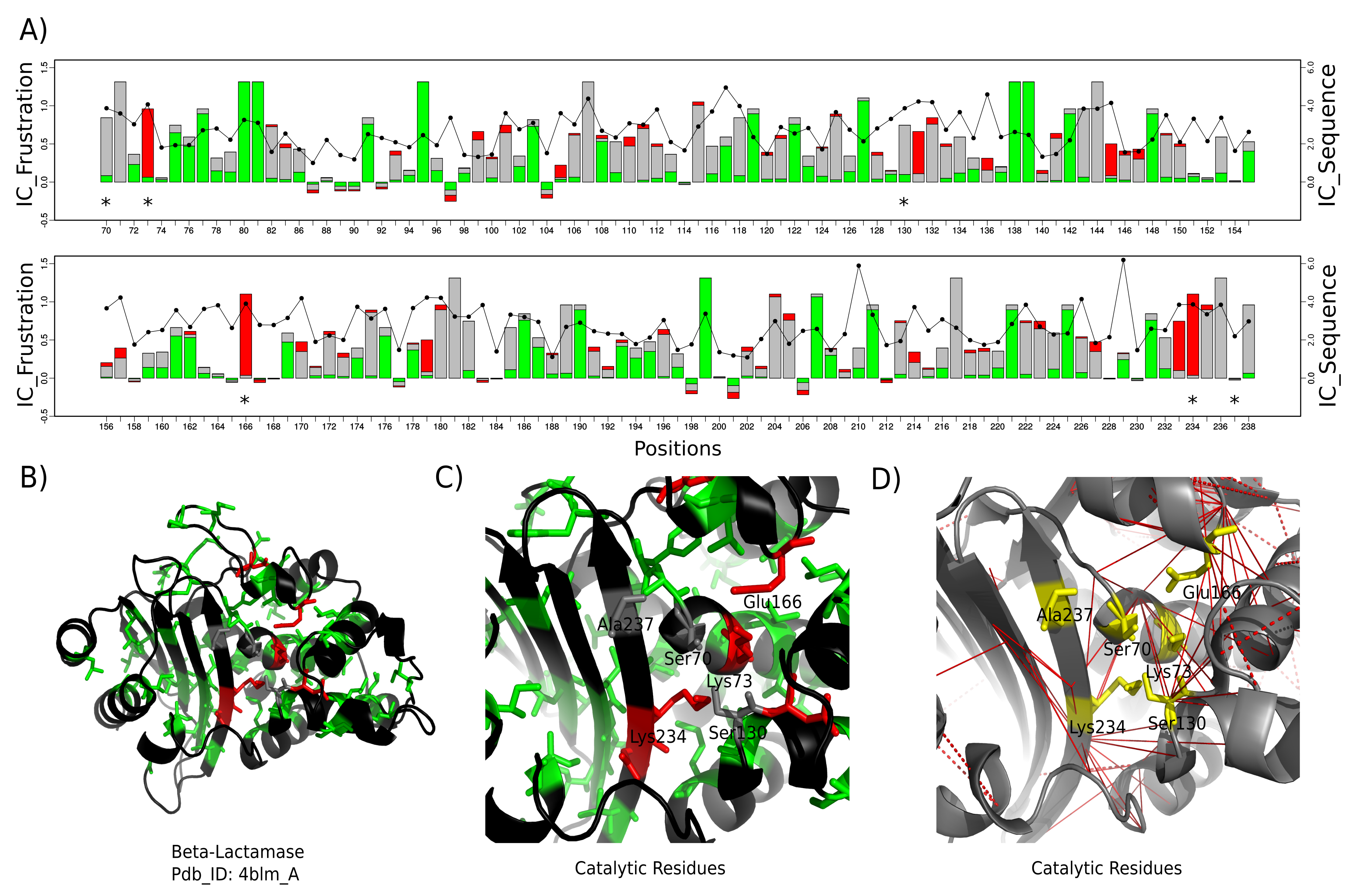
Beta-Lactamases Class A Family has five catalytic residues, Ser70, Lys73, Ser130, Glu166, Lys234, and Ala237. For this analysis we performed a multiple sequence alignment (MSA) of 31 homologs. We used a non redundant set of experimental structures that correspond to the 31 sequences in the MSA, one for each sequence. Using FrustraEvo we calculate the information content of frustration states (FrstIC) for position in the MSA for all of the structures of members of a family. We have found that Lys73, Glu166, and Lys234 are 'highly frustrated' and conserved (IC > 0.5). Ser70 and Ser130 are 'neutral' and conserved and Ala237 is not conserved (IC < 0.5). We also observe five non-catalytic residues (Tyr105, Asp131, Pro145, Asp179 and Asp233) that are conserved and 'highly frustrated'. These residues are involved in different functionalities, for example Tyr105 is involved in substrate recognition and binding. Mutations in Asp179 affect the resistance of beta-lactamases to different antibiotics. We also detected some positions that display high sequence variability, the local frustration signature is retained. For some other positions, the frustration state is sensitive to changes in sequence.