PLPro Family (SARSCov2 Protease)
This is a case study that was analyzed in one of our articles where we generalize the use of FrustraEvo to study protein families stability and functional patterns: Freiberger et al Nature Comms 2023. We analysed the differential frustration patterns among the different PLPro subfamilies within the coronaviruses phylogeny. In what follows we repdocue some of the analysis of the article.
In the following link you can find a FrustraEvo reproduction of the SARSCov2 PLPro resundant set of structures that is part of this example (first row as shown in the summary figure below):
FrustraEvo job results
Biological Interpretation:
When comparing protein families across different Coronaviruses we also observed a large variability between different subfamilies. Some positions in some proteins are conserved across the entire phylogenetic tree, while others have energetic signatures that are specific to some subfamilies like the Sarbecoviruses or to specific viruses like the SARS-CoV-2.
PLPro catalyzes the proteolysis of the viral polyproteins. Moreover, PLPro interacts with at least two host proteins, ubiquitin-like interferon-stimulated gene 15 protein (ISG15) and ubiquitin (Ub), to evade or at least hamper the host immune response31. SARS-CoV-2 PLPro homologous proteins were automatically divided into 4 subfamilies that reflect the Betacoronavirus subgenera classification, i.e., Sarbecovirus (n = 31), Nobecovirus (n = 11), Merbecovirus (n = 35) and Embecovirus (n = 45). Additionally, we have manually analyzed a fifth group that only contains experimental SARS-CoV-2 PLPro structures (n = 29) to quantify frustration conservation specific to this virus. We compared frustration conservation between the 4 PLPro subfamilies to disclose functional diversity related to differential infectivity or virulence (see Fig.). At the catalytic site, the SDP Trp106, which facilitates catalysis by stabilizing the catalytic triad, is conserved both in sequence and in its highly frustrated state only in the Sarbecoviruses group. In that same position, Merbecoviruses have a conserved Leu that is not energetically conserved, which is reported to make catalysis less efficient. When Leu 106 is replaced by a Trp in MERS, catalysis is enhanced, suggesting that increased local frustration may be related to the improvement of the catalytic function. In contrast, the catalytic residue Cys111 is minimally frustrated and conserved in all subfamilies, reflecting the functional importance of local stability at that position to the full phylogeny. In the SARS-CoV-2 set of structures, this position appears neutral, due to the occurrence of a subgroup that contains the Cys111Ser mutation, which introduces local instabilities. Likewise, the four cysteines (Cys189, Cys192, Cys224, and Cys226) that coordinate the binding of an ion of Zinc, indispensable for the functioning of the protein, are all minimally frustrated and conserved in most of the coronavirus subfamilies, suggesting a strong stability requirement in that region (see Fig.).
Additionally, the PLPro binding sites to ISG15 and Ub host proteins (S1 and S2, respectively), are differentially conserved between the four Betacoronavirus subfamilies (see Fig.). The SARS-CoV-2 S1 site contains more highly frustrated residues while the S2 site contains more minimally frustrated residues than the other viruses. This could explain the differential preference that PLPpro has for binding to ISG15 or Ub in SARS-CoV and SARS-CoV-234. For instance, some positions within the S1 region are highly frustrated only at the SARS-CoV-2 level. Positions 225 and 232 (SARS-CoV-2 numbering) (see Fig.) correspond to neutral Val and Gln in Sarbecovirus but to highly frustrated Thr and Lys in SARS-CoV-2. It has been shown that these changes affect Ub association, explaining the differential activity on Ub substrates but not on ISG1531. Thr225 is only present in SARS-CoV-2 and in RaTG13, the latter being a likely bat progenitor of the COVID-19 virus35. Moreover, the bat-derived viral strains, Rc-o319, and bat-SL-CoVZXC21, contain a Met in that position that is even more frustrated. This may point to a position of concern for novel human-infecting variants that could acquire this change of identity. Lys232 is unique to SARS-CoV-2 within the Sarbecovirus family, suggesting a recent gain of function event.
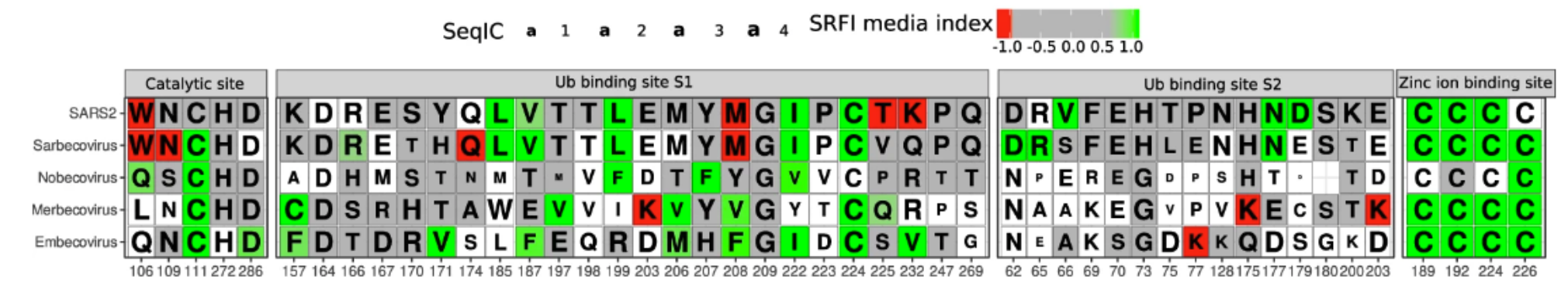
MSFA showing FrustraEvo results for selected functional domains in PLPro. Cells that are colored correspond to FrustIC > 0.5, while white cells mean that FrustIC ≤ 0.5. Color of the cells represents the median SRFI value computed with FrustratometeR (see methods for frustration states definitions). The amino acid identities correspond to the consensus sequence, and the size of the letter is proportional to SeqIC.